This topic takes on average 55 minutes to read.
There are a number of interactive features in this resource:
 Biology
Biology
 Human biology
Human biology
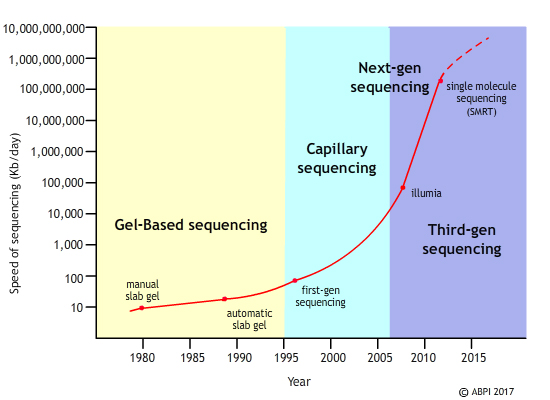
The Human Genome Project took 13 years to sequence the first human genome at a cost of almost three billion dollars. Now a human genome can be sequenced in a matter of days and a cost of a few thousand dollars. The speed and accuracy of the process are increasing all the time while the cost is falling. Since the first human genome was sequenced using the Sanger method of analysis several different technologies have had an influence on the way scientists analyse genomes.
Next-generation sequencing
Next-generation sequencing was introduced in 2007. The big breakthrough with this technique is that millions of DNA fragments can be sequenced at the same time. As a reflection of this, it is sometimes known as massively parallel sequencing. The preparation of the material is faster and easier – some next-generation sequencing machines can produce more than five complete human genomes in under a week. It is a very flexible technology – but it is already being overtaken by the next development in the process.

There are a number of third-generation technologies in development or in use already. They offer the opportunity for even faster, even more accurate and even cheaper genome analysis. One of these new techniques is known as Single-Molecule Sequencing in Real Time (SMRT). Here, a single stranded DNA molecule is sequenced as a DNA polymerase moves down the strand, attaching complementary fluorescently-labelled bases as it goes. The colour of each fluorescent tag is recorded to identify the base before the tag is removed and then the next base added. It gives scientists a detailed insight into the building of a new DNA molecule and an accurate record of the bases in the DNA strand. It is incredibly accurate and sensitive, opening the doors to a whole new way of using the human genome. At the moment it cannot be used to read a human DNA sequence but that is only a matter of time. Once third-generation machines can sequence human genomes, they will be able to complete the entire sequence in around an hour. This starts to open the door to real personalised medicine, with patient genomes available to hospitals and even, in the future, to GPs.
Genome sequencing generates enormous amounts of data. It is one of many areas where computational biology and bioinformatics is becoming ever more important. Making sense of the mass of data generated by the new sequencing technologies requires software and computing tools including appropriate algorithms, statistical tests and mathematical models. Whole new areas of understanding are opening up as rapid data generation is matched by new ways of using computers to analyse and present the data, so we can find and use the patterns which emerge.